The first thing most people do with FeatureFinder is setup a profile and add their data files. But, if your files aren’t in the “one-trial-per-file,” text-file format that FeatureFinder expects, you’ll want to do a couple things first: 1) convert your data files to text, and 2) split your data files by trial. This tutorial will show you how to do this—and give you all the code you need! We’ll start by getting your data into text format…
Exporting your data
Your data may already be in text format—i.e., it has a .txt filename and opens in WordPad/TextEdit/etc.—, but if not, there’s almost always an option in your data collection or analysis program to export data to text. Sometimes this will be in the “Save As” window, like with BIOPAC AcqKnowledge, or it could be elsewhere in the program’s menus (e.g., under “Export…”).
If you’re using BIOPAC’s AcqKnowledge program, here’s what to do:
- Open up your
.acqdata file in AcqKnowledge - Click “Save As” under the “File” menu
- Select “Text” under the “Save as type” drop-down
- Click the Options button and ensure the following:
- “Include header” is checked
- “Selected section only” is unchecked
- “Horizontal scale values” is unchecked
Splitting your data
Downloads for this tutorial:
NOTE: The two m-files must be unzipped prior to use.
Now that your data file is in text format, we can split it up by trial. We’ll look at two scenarios: trials with equal spacing (say, starting at 5 s, 10 s, 15 s, etc.) and trials with unequal spacing (say: 2, 10, 14, 22, etc.). To make sure we’re on the same page throughout this tutorial, we’re going to use the example files EqualSpacing.txt and UnequalSpacing.txt. Feel free to download them and follow along!
So, just to review, we’re taking a file that has lots of trials:
UnequalSpacing.txt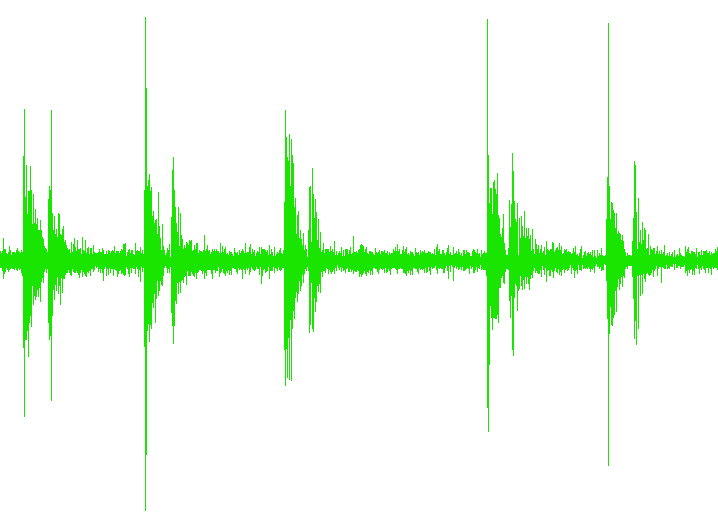
…and separating the trials into separate files, like this:
...Trial1.txt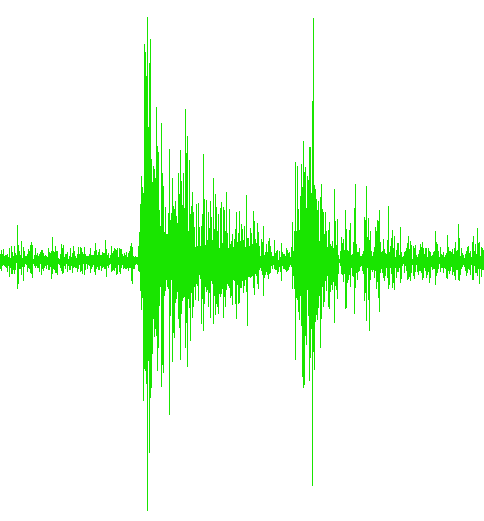
...Trial2.txt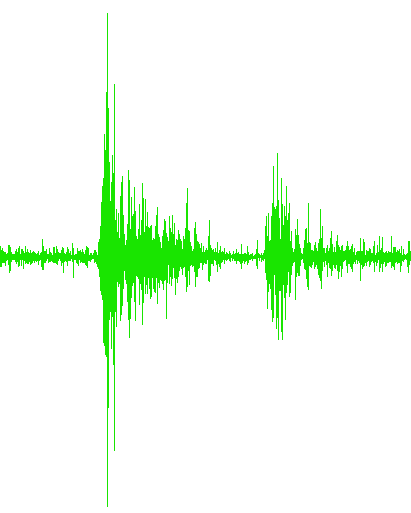
...Trial3.txt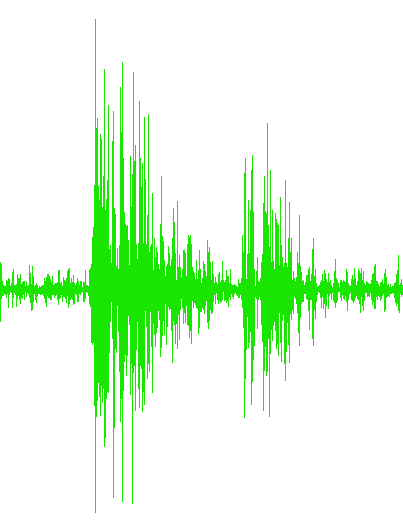
...Trial4.txt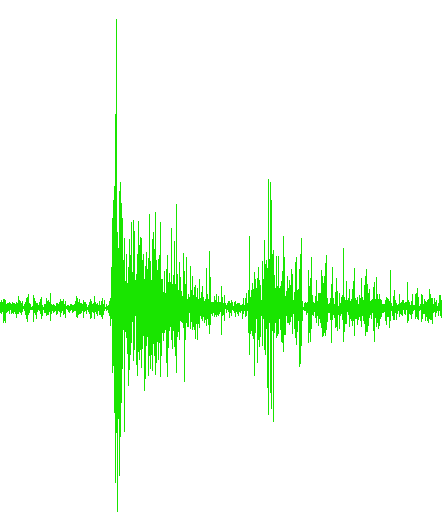
...Trial5.txt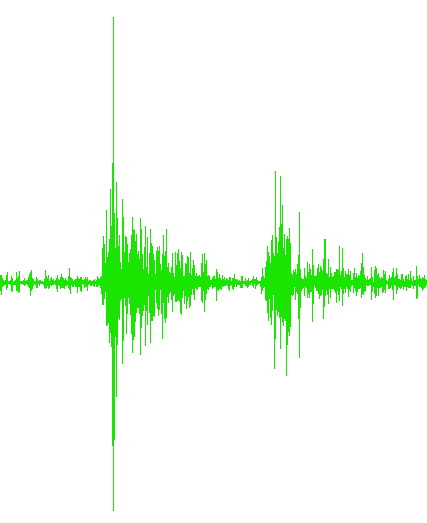
Here’s how to do it:
- Make sure you know i) the sampling rate of your data, ii) the start time of each trial within your data file, and iii) the length of each trial.
- For our example data file,
EqualSpacing.txt, the sampling rate is 1000 Hz, the trials start at 2, 8, 14 and 20 s, and they are all 6 s long. - For
UnequalSpacing.txt, the sampling rate is also 1000 Hz, but the trials start at 0, 6, 13, 23, and 29 s. All trials are again 6 s. - Download and unzip the MATLAB files
SplitData.mandUtil_GetFileInfo.mto the same directory as your data file(s). - Set MATLAB’s “Current Folder” to the directory containing the downloaded files and your data.
- In the “Command Window,” type the following command (all on one line):
SplitData('Filename', [T1Start T1Length; T2Start T2Length; etc.], SamplingRate);replacing the words above with their actual values, and press Enter. Note the single quotation marks around the filename and the semicolons between each pair of trial start/length values.
For EqualSpacing.txt, we’d type the following:
SplitData('EqualSpacing.txt', [2 6; 8 6; 14 6; 20 6], 1000)For UnequalSpacing.txt, we’d type something similar:
SplitData('UnequalSpacing.txt', [0 6; 6 6; 13 6; 23 6; 29 6], 1000)And you’re done! Take a look in your data directory and you’ll see all of your new files. If you’ve been following along with the example data, then the new files will have the names Equal_Trial1.txt, Unequal_Trial3.txt, and so on.
If you have any trouble splitting your data, please drop us a note and we’d be happy to offer some advice. And, once your data is in text form and all split up, you’ll be ready to start using FeatureFinder! Check out our written and video tutorials to get started!


hi
i am need paper and larg database EMG(sample data) for thesis.
please help me.
thanks.
Yup, it’s cool. I had it corrupt some of my files It silmpy wouldn’t convert them correctly. Don’t remember what the problem was though Though, it had something to do with files, and not with iconv. It worked with jedit
Hi Ali, you can download our sample EMG data by clicking on the “Download” link in the menu bar, and then “Sample Data,” or by going straight to https://www.featurefinder.ca/sample-data/. Hope that helps!
Hi Zelli, glad it worked out in the end, but I’d be interested to hear more about what happened. Could you drop us a note to give us more information? (Click “Community” in the menu bar, and then “Contact Us”)
plz i want to have more information about the EMG data
(like the time of the to catching the object, ….etc)
Hi Zizou, thanks for the comment. The files were setup so that the object was dropped two seconds in. You can find some more details about the EMG data collection at https://www.featurefinder.ca/sample-data/.